Subject:
General Approach to Evaluating the Utility of Genetic Panels
Description:
_______________________________________________________________________________________
IMPORTANT NOTE:
The purpose of this policy is to provide general information applicable to the administration of health benefits that Horizon Blue Cross Blue Shield of New Jersey and Horizon Healthcare of New Jersey, Inc. (collectively “Horizon BCBSNJ”) insures or administers. If the member’s contract benefits differ from the medical policy, the contract prevails. Although a service, supply or procedure may be medically necessary, it may be subject to limitations and/or exclusions under a member’s benefit plan. If a service, supply or procedure is not covered and the member proceeds to obtain the service, supply or procedure, the member may be responsible for the cost. Decisions regarding treatment and treatment plans are the responsibility of the physician. This policy is not intended to direct the course of clinical care a physician provides to a member, and it does not replace a physician’s independent professional clinical judgment or duty to exercise special knowledge and skill in the treatment of Horizon BCBSNJ members. Horizon BCBSNJ is not responsible for, does not provide, and does not hold itself out as a provider of medical care. The physician remains responsible for the quality and type of health care services provided to a Horizon BCBSNJ member.
Horizon BCBSNJ medical policies do not constitute medical advice, authorization, certification, approval, explanation of benefits, offer of coverage, contract or guarantee of payment.
__________________________________________________________________________________________________________________________
Genetic panel testing offers potential advantages and disadvantages compared with direct sequence analysis. This conceptual framework outlines a structure for evaluating the utility of genetic panels, by classifying them into clinically relevant categories and developing criteria for evaluating panels in each category.
Genetic panels using next-generation technology or chromosomal microarray analysis are available for many clinical conditions. The major advantage of panels is the ability to analyze many genes simultaneously, potentially improving the breadth and efficiency of the genetic workup. A potential disadvantage of panels is that they provide a large of amount of ancillary information whose significance may be uncertain. Limited published evidence has reported that the analytic validity of panels approaches that of direct sequencing. The clinical validity and clinical utility of panels are condition-specific. The clinical validity of panels will reflect the clinical validity of the underlying individual variants. The clinical utility of panels will depend on the context in which they are used, ie, whether the advantages of panel testing outweigh the disadvantages for the specific condition under consideration.
Panels can be classified into categories based on their intended use and composition. For each category of panels, specific criteria can be used to evaluate medical necessity. When all criteria for a given category are met, that panel may be considered medically necessary.
| Populations | Interventions | Comparators | Outcomes |
Individuals:
- Who are symptomatic with a suspected genetically associated disease
| Interventions of interest are:
- Genetic panel testing for a suspected genetically associated disorder
| Comparators of interest are:
- Standard clinical management without genetic panel testing
| Relevant outcomes include:
- Test accuracy
- Test validity
- Disease-specific survival
- Overall survival
- Change in disease status
- Morbid events
- Functional outcomes
- Changes in reproductive decision making
|
Individuals:
- Who are asymptomatic and have a close relative diagnosed with a genetically associated disease
| Interventions of interest are:
- Genetic panel testing for a genetically associated disorder
| Comparators of interest are:
- Standard clinical management without genetic panel testing
| Relevant outcomes include:
- Test accuracy
- Test validity
- Disease-specific survival
- Overall survival
- Change in disease status
- Morbid events
- Functional outcomes
- Changes in reproductive decision making
|
BACKGROUND
This conceptual framework applies if there is not a separate policy that outlines specific criteria for testing. If a separate policy does exist, then the criteria for medical necessity therein supersede the guidelines herein.
Context
The purpose of this policy is to provide a framework for evaluating the utility of genetic panels that use newer genetic testing methodologies. In providing a framework for evaluating genetic panels, this review will not attempt to determine the clinical utility of genetic testing for specific disorders per se. For most situations, this will mean that at least 1 variant in the panel has already been determined to have clinical utility and that clinical indications for testing are established. Once the clinical utility for at least one of the variants included in the panel has been established, then the focus is on whether the use of a panel is a reasonable alternative to individual tests.
Genetic Panel TESTING
A genetic panel will be defined as a test that simultaneously evaluates multiple genes, as opposed to sequential testing of individual genes. This includes panels performed by next-generation sequencing (NGS), massive parallel sequencing, and chromosomal microarray analysis. The definition of a panel will not include panels that report on gene expression profiling, which generally do not directly evaluate genetic variants.
New Sequencing Technologies
New genetic technology, such as NGS and chromosomal microarray, has led to the ability to examine many genes simultaneously.1, This in turn has resulted in a proliferation of genetic panels. Panels using next-generation technology are currently widely available, covering a broad range of conditions related to inherited disorders, cancer, and reproductive testing.2,3,4, These panels are intuitively attractive to use in clinical care because they can analyze multiple genes more quickly and may lead to greater efficiency in the workup of genetic disorders. It is also possible that newer technology can be performed more cheaply than direct sequencing, although this may not be true in all cases.
Newer sequencing techniques were initially associated with higher error rates than direct sequencing.5 While there are limited published data directly comparing the accuracy of NGS with direct sequencing, several publications have reported that the concordance between NGS and Sanger sequencing is greater than 99% for cancer susceptibility testing,6 inherited disorders,7 and hereditary hearing loss.8, Another potential pitfall is the easy availability of a multitude of genetic information, much of which has uncertain clinical consequences. Variants of uncertain significance are found commonly and in greater numbers with NGS than with direct sequencing.9,10,
The intended use for these panels is variable, For example, for the diagnosis of hereditary disorders, a clinical diagnosis may be already established, and genetic testing is performed to determine whether this is a hereditary condition, and/or to determine the specific variant present. In other cases, there is a clinical syndrome (phenotype) with a broad number of potential diagnoses, and genetic testing is used to make a specific diagnosis. For cancer panels, there are also different intended uses. Some panels may be intended to determine whether a known cancer is part of a hereditary cancer syndrome. Other panels may include somatic variants in a tumor biopsy specimen that may help identify a cancer type or subtype and/or help select the best treatment.
There is no standardization to the makeup of genetic panels. Panel composition is variable, and different commercial products for the same condition may test a different set of genes. The makeup of the panels is determined by the specific lab that developed the test. Also, the composition of any individual panel is likely to change over time, as new variants are discovered and added to existing panels.
Despite the variability in the intended use and composition of panels, there are a finite number of broad panel types that can be identified and categorized. Once categorized, specific criteria on the utility of the panel can be developed for each category. One difficulty with this approach is that the distinction between the different categories, and the distinction between the intended uses of the panels, may not be clear. Some panels will have features or intended uses that overlap among the different categories.
To determine the criteria used for evaluating panels, the policy will first classify panels into a number of clinically relevant categories, according to their intended use. Then, for each category, criteria will be proposed that can be applied to tests within that category. Because our goal is to outline a general approach to testing, we will not evaluate individual panels; rather, we will supply examples of genetic panels in each category to assist Plans in classifying the individual panels.
Regulatory Status
Clinical laboratories may develop and validate tests in-house and market them as a laboratory service; laboratory-developed tests must meet the general regulatory standards of the Clinical Laboratory Improvement Amendments. Laboratories that offer laboratory-developed tests must be licensed by the Clinical Laboratory Improvement Amendments for high-complexity testing. To date, the U.S. Food and Drug Administration has chosen not to require any regulatory review of this test.
An exhaustive list of commercially available panel tests is impractical. For example, the EGL Genetics offers 243 different genetic panels, of a total of 929 molecular genetics tests.11, Table 1 provides a sample of panels that use NGS or chromosomal microarray technologies.
Table 1. Panels Using Next-Generation Sequencing or Chromosomal Microarray Analysis (as of December 2017)
Test Name | Laboratory |
| Agammaglobulinemia Panel | ARUP Laboratories |
| Ashkenazi Jewish Diseases Panel | ARUP Laboratories |
| Mitochondrial Disorders Panel | ARUP Laboratories |
| Amyotrophic Lateral Sclerosis Pane | ARUP Laboratories |
| Aortopathy Panel | ARUP Laboratories |
| Autism Panel | ARUP Laboratories |
| Brugada Syndrome Panel | ARUP Laboratories |
| Vascular Malformation Syndromes | ARUP Laboratories |
| Retinitis Pigmentosa/Leber Congenital Amaurosis Panel | ARUP Laboratories |
| Cardiomyopathy and Arrhythmia Panel | ARUP Laboratories |
| Periodic Fever Syndromes Panel | ARUP Laboratories |
| Arrhythmias Sequencing Panel | EGL Genetics |
| Arrhythmias Deletion/Duplication Panel | EGL Genetics |
| Autism Spectrum Disorders | EGL Genetics |
| Cardiomyopathy Panel | EGL Genetics |
| Ciliopathies Panel | EGL Genetics |
| Congenital Glycosylation Disorders | EGL Genetics |
| ACOG/ACMG Carrier Screen Targeted Mutation Panel | EGL Genetics |
| Epilepsy | EGL Genetics |
| Eye Disorders | EGL Genetics |
| Neuromuscular Disorders | EGL Genetics |
| Noonan Syndrome and Related Disorders | EGL Genetics |
| Short Stature Panel | EGL Genetics |
| Sudden Cardiac Arrest Panel | EGL Genetics |
| X-linked Intellectual Disability | EGL Genetics |
| CancerNext™ | Ambry Genetics |
| BreastNext™ | Ambry Genetics |
| ColoNext™ | Ambry Genetics |
| OvaNext™ | Ambry Genetics |
| RhythmNext® | Ambry Genetics |
| X-linked Intellectual Disability | Ambry Genetics |
| TAADNext® | Ambry Genetics |
| Cobalamin Metabolism Comprehensive Panel | Baylor College of Medicine |
| Progressive External Ophthalmoplegia Panel | Baylor College of Medicine |
| CoQ10 Comprehensive Panel | Baylor College of Medicine |
| Usher Syndrome Panel | Baylor College of Medicine |
| Retinitis Pigmentosa Panel | Baylor College of Medicine |
| Pyruvate Dehydrogenase Deficiency and Mitochondrial Respiratory Chain Complex V Deficiency Panel | Baylor College of Medicine |
| Myopathy/Rhabdomyolysis Panel | Baylor College of Medicine |
| Mitochondrial Disorders Panel | Baylor College of Medicine |
| Low Bone Mass Panel | Baylor College of Medicine |
| Glycogen Storage Disorders Panel | Baylor College of Medicine |
| Leigh Disease Panel | Medical Neurogenetics |
| Pan Cardiomyopathy Panel | Partners Healthcare |
| Isolated Non-syndromic Congenital Heart Defects Panel | Partners Healthcare |
| Noonan Spectrum Panel | Partners Healthcare |
| Usher Syndrome Panel | Partners Healthcare |
| Hereditary Colon Cancer Syndromes | Mayo Medical Laboratories |
| Hypertrophic Cardiomyopathy Panel | Mayo Medical Laboratories |
| Dilated Cardiomyopathy Panel | Mayo Medical Laboratories |
| Arrhythmogenic Right Ventricular Cardiomyopathy Panel | Mayo Medical Laboratories |
| Noonan Syndrome Panel | Mayo Medical Laboratories |
| Marfan Syndrome Panel | Mayo Medical Laboratories |
| Long QT Syndrome | Mayo Medical Laboratories |
| Brugada Syndrome | Mayo Medical Laboratories |
| Signature Prenatal Microarray | Signature Genomics |
| Counsyl™ Panel | Counsyl Genomics |
| GoodStart Select™ | GoodStart Genetics |
Related Policies
- General Approach to Genetic Testing (Policy #082 in the Pathology Section)
- Genetic Cancer Susceptibility Panels Using Next Generation Sequencing (Policy #084 in the Pathology Section)
- Expanded Molecular Panel Testing of Cancers to Identify Targeted Therapies (Policy #105 in the Pathology Section)
Policy:
(NOTE: For services provided August 1, 2017 and after, Horizon Blue Cross Blue Shield of New Jersey collaborates with eviCore healthcare to conduct Medical Necessity Determination for certain molecular and genomic testing services for members enrolled in Horizon BCBSNJ fully insured products as well as Administrative Services Only (ASO) accounts that have elected to participate in the Molecular and Genomic Testing Program (“the Program”). Beginning August 1, 2017, the criteria and guidelines included in this policy apply to members enrolled in plans that have NOT elected to participate in the Program.
To access guidelines that apply for services provided August 1, 2017 and after to members enrolled in plans that HAVE elected to participate in the Program, please visit www.evicore.com/healthplan/Horizon_Lab.
For Medicare Advantage, please refer to the Medicare Coverage Section below for coverage guidance.
This policy applies only if there is not a separate medical policy that outlines specific criteria for testing. If a separate medical policy does exist, then the criteria for medical necessity in that policy supersede the guidelines in this policy.)
I. Genetic panels that use next-generation sequencing or chromosomal microarray analysis, and are classified in one of the categories below, is considered medically necessary when all criteria are met for each category, as outlined below:
- Panels for hereditary or genetic conditions
o Diagnostic testing of an individual’s germline to benefit the individual
o Testing of an asymptomatic individual to determine future risk of disease
- Cancer panels
o Testing of an asymptomatic individual to determine future risk of cancer
o Testing cancer cells from an individual to benefit the individual by identifying targeted treatment
- Reproductive panels
o Preconception testing
- Carrier testing of the parent(s)
o Prenatal testing
- Carrier testing of the parent(s)
- In utero testing of a fetus, including testing for aneuploidy or familial variants
o Preimplantation genetic testing.
II. Genetic panels that use next-generation sequencing or chromosomal microarray that do not meet the criteria for a specific category are considered investigational.
(NOTE: Criteria for Evaluating Genetic Panels
The following are criteria that can be applied to evaluating genetic panels, with an explanation of the way the criteria are to be defined and applied. Not all criteria will apply to all panels. Appendix Table 2 and Appendix Figures 1 through 4 list the specific criteria that should be used for each category.
Test Is Performed in a Clinical Laboratory Improvement Amendments‒Licensed Lab
· Testing is performed in a laboratory licensed under Clinical Laboratory Improvement Amendments for high-complexity testing. This requires delivery of a reproducible set of called, quality-filtered variants from the sequencing platform.
· These calculations should occur before variant annotation, filtering, and manual interpretation for patient diagnosis.
Technical Reliability of Panels Approaches That of Direct Sequencing
· The technical reliability for detecting individual variants, compared with the criterion standard of conventional direct Sanger sequencing, is reported.
o The testing methods are described, and the overall analytic validity for that type of testing is defined.
· Any decrease in analytic sensitivity and specificity is not large enough to result in a clinically meaningful difference in diagnostic accuracy (clinically valid).
All individual components of the panel have demonstrated they are clinically useful for the condition being evaluated OR the implications and consequences of test results that have not demonstrated clinical utility are clear, AND there is no potential for incidental findings to cause harm.
· For each panel, if each variant in the panel would be indicated for at least some patients with the condition, then this criterion is met.
o If there are individual variants that do not have clinical utility, then the potential to cause harm might occur.
· For incidental findings, the potential for harm may be due to:
o Incorrect diagnosis due to false-positive or false-negative results
§ False-positive: Unnecessary treatment that may have adverse events
§ False-negative: Effective treatment not provided
o Incorrect risk assessment
§ Unnecessary surveillance tests may lead to further confirmatory tests that may be invasive
§ Effective surveillance or screening not provided to patients at-risk
§ Incorrect decision made on reproductive decision making
· Alteration made in reproductive planning that would not have been made with correct information
· No alteration made in reproductive planning, where alteration would have been made with correct information
Panel Testing Offers Substantial Improvement in Efficiency vs Sequential Analysis of Individual Genes
· The composition of the panel is sufficiently complex such that next-generation sequencing or chromosomal microarray analysis is expected to offer considerable advantages. The complexity of testing can be judged by:
o The number of genes tested.
o The size of the genes tested.
o The heterogeneity of the genes tested.
The Impact of Ancillary Information Is Well-Defined
· If a panel contains both variants that are medically necessary and variants that are investigational (or not medically necessary), the impact of results for investigational (or not medically necessary) variants is considered, taking into account the following possibilities:
o The information may be ignored (no further impact).
o The information may result in further testing or changes in management:
§ Positive impact
§ Negative impact
· It is more likely that the results of tests that are not medically necessary cause a negative, rather than a positive, impact on the patient. This is because additional tests and management changes that follow are not evidence-based and because additional testing and treatment generally involve risks.
Decision Making Based on Genetic Results Is Well-Defined
· Results of the genetic testing will lead to changes in diagnosis and/or treatment.
· The potential changes in treatment are defined prior to testing and accord with the current standard of care.
· Changes in diagnosis or management are associated with improvements in health outcomes.
· For prenatal and preconception testing:
o Alterations in reproductive decision making are expected, depending on the results of testing.
Testing Yield Is Acceptable for the Target Population
· The number of individuals who are found to have a pathogenic variant, in relation to the total number of individuals tested, is reasonable given the underlying prevalence and severity of the disorder, and the specific population that is being tested.
o It is not possible to set an absolute threshold for acceptable yield across different clinical situations. Some guidance can be given from clinical precedence as follows:
§ For diagnosis of hereditary disorders, genetic testing is generally performed when signs and symptoms of the disease are present, including family history. The likelihood of a positive genetic test depends on the accuracy of the signs and symptoms (pretest probability of disorder), and the clinical sensitivity of genetic testing. For disorders such as testing for congenital long QT syndrome and Duchenne muscular dystrophy, the likelihood of a positive result in patients with signs and symptoms of the disease is greater than 10%.
§ For cancer susceptibility, testing is recommended for genetic abnormalities such as the BRCA gene and Lynch syndrome when the likelihood of a positive result is in the range of 2% to 10%.
§ For a clinical syndrome that has multiple underlying etiologies, such as developmental delay in children, chromosomal microarray analysis is recommended when the likelihood of a positive result is in the 5% to 20% range.
· There is an increase in yield over alternative methods of diagnosis, and this increase is clinically significant.
Other Issues to Consider
· Most tests will not, and possibly should not, be ordered by generalists.
o Guidance for providers is appropriate on the expertise necessary to ensure that test ordering is done optimally.
· Many tests, particularly those for inherited disorders, should be accompanied by patient counseling, preferably by certified genetic counselors.
o Counseling may be needed both before and after testing, depending on the specific condition being tested.)
Medicare Coverage:
Per NCD 90.2 effective March 16, 2018, NGS is covered if the test is performed in a CLIA-certified laboratory, when ordered by a treating physician and when all of the following criteria are met:
· The Individual has either recurrent, relapsed, refractory, metastatic, or advanced stages III or IV cancer; and
· The Individual has either not been previously tested using the same NGS test for the same primary diagnosis of cancer or repeat testing using the same NGS test only when a new primary cancer diagnosis is made by the treating physician; and
· The Individual has decided to seek further cancer treatment (e.g., therapeutic chemotherapy).
The diagnostic laboratory test using NGS must have:
· Food & Drug Administration (FDA) approval or clearance as a companion in vitro diagnostic; and,
· an FDA-approved or -cleared indication for use in that patient’s cancer; and,
· results provided to the treating physician for management of the patient using a report template to specify treatment options.
For additional information and eligibility for NGS, refer to National Coverage Determination (NCD) for Next Generation Sequencing (NGS) (90.2). Available at: https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=290.
On 1/27/20, CMS issued Decision Memo for Next Generation Sequencing (NGS) for Medicare Beneficiaries with Advanced Cancer (CAG-00450R) with additional coverage for next generation sequencing. CMS also provided that Medicare Administrative Contractors (MACs) may determine coverage of Next Generation Sequencing (NGS) as a diagnostic laboratory test when performed in a CLIA-certified laboratory, when ordered by a treating physician, when results are provided to the treating physician for management of the individual. For additional information and eligibility, refer to Decision Memo for Next Generation Sequencing (NGS) for Medicare Beneficiaries with Advanced Cancer (CAG-00450R). Available at:https://www.cms.gov/medicare-coverage-database/indexes/nca-open-and-closed-index.aspx?type=Closed.
Also, please refer to:
Policy Guidelines: (Information to guide medical necessity determination based on the criteria contained within the policy statements above.)
Genetics Nomenclature Update
The Human Genome Variation Society nomenclature is used to report information on variants found in DNA and serves as an international standard in DNA diagnostics. It is being implemented for genetic testing medical policyupdates starting in 2017 (see Table PG1). The Society’s nomenclature is recommended by the Human Variome Project, the HUman Genome Organization, and by the Human Genome Variation Society itself.
The American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines for interpretation of sequence variants represent expert opinion from both organizations, in addition to the College of American Pathologists. These recommendations primarily apply to genetic tests used in clinical laboratories, including genotyping, single genes, panels, exomes, and genomes. Table PG2 shows the recommended standard terminology—“pathogenic,” “likely pathogenic,” “uncertain significance,” “likely benign,” and “benign”—to describe variants identified that cause Mendelian disorders.
Table PG1. Nomenclature to Report on Variants Found in DNA
Previous | Updated | Definition |
| Mutation | Disease-associated variant | Disease-associated change in the DNA sequence |
 | Variant | Change in the DNA sequence |
 | Familial variant | Disease-associated variant identified in a proband for use in subsequent targeted genetic testing in first-degree relatives |
Table PG2. ACMG-AMP Standards and Guidelines for Variant Classification
Variant Classification | Definition |
| Pathogenic | Disease-causing change in the DNA sequence |
| Likely pathogenic | Likely disease-causing change in the DNA sequence |
| Variant of uncertain significance | Change in DNA sequence with uncertain effects on disease |
| Likely benign | Likely benign change in the DNA sequence |
| Benign | Benign change in the DNA sequence |
American College of Medical Genetics and Genomics; AMP: Association for Molecular Pathology.
Genetic Counseling
Experts recommend formal genetic counseling for patients who are at-risk for inherited disorders and who wish to undergo genetic testing. Interpreting the results of genetic tests and understanding risk factors can be difficult for some patients; genetic counseling helps individuals understand the impact of genetic testing, including the possible effects the test results could have on the individual or their family members. It should be noted that genetic counseling may alter the utilization of genetic testing substantially and may reduce inappropriate testing; further, genetic counseling should be performed by an individual with experience and expertise in genetic medicine and genetic testing methods.
[RATIONALE: This conceptual framework was created in June 2013 and has been updated regularly with searches of the MEDLINE database. The most recent literature update was performedthrough November 7, 2017.
Types of Panel Testing
There are numerous types of panel testing, because in theory a panel may be substituted for individual variant testing in any situation where more than 1 gene is being examined. Commercially available panels fall largely into several categories, which we classify using the BCBSA categories of genetic testing (see Appendix Table 1).
We have classified genetic panels into 3 major categories: panels for genetic and hereditary conditions, cancer panels, and reproductive panels. Within these categories, we created subcategories by the intended use of the panels.
Panels for Genetic or Hereditary Conditions
Panels for genetic or hereditary conditions are generally single-gene disorders, which are inherited in Mendelian fashion. They are defined by a characteristic phenotype, which may characterize a specific disease or represent a syndrome that encompasses multiple underlying diseases.
The intended use of these panels may be for:
· Diagnostic testing of an individual’s germline to benefit the individual. To confirm a suspected diagnosis in patients with signs and/or symptoms of the condition; or to identify a causative etiology for a clinical syndrome, for which there are multiple possible underlying conditions.
· Testing an asymptomatic individual to determine future risk of disease.
There are several variations of panels for use in diagnosis or risk assessment of genetic or hereditary conditions. For our purposes, panels will be divided into the following types:
· Panels containing variants associated with a single condition. These panels generally include all known pathogenic variantsfor a defined disease and do not include variantsassociated with other diseases. An example of such a panel would be one that includes pathogenic variants for hypertrophic cardiomyopathy but does not include variants associated with other cardiovascular disorders. These panels can be used for diagnostic or risk assessment purposes.
· Panels containing variants associated with multiple related conditions. These panels include all known pathogenic variantsfor a defined disease and variants associated with other related disorders. An example of such a panel would be a pan cardiomyopathy panel that includes pathogenic variantsfor hypertrophic cardiomyopathy and other types of cardiomyopathy (eg, dilated cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy). These panels can be used for diagnostic or risk assessment purposes.
· Panels containing variants for clinical syndromes associated with multiple distinct conditions. These panels include variants associated with multiple potential disease states that define a particular clinical syndrome. In general, a specific diagnosis cannot be made without genetic testing, and genetic testing can identify one among several underlying disease states that manifest as a clinical syndrome. An example of this type of panel is one for intellectual disability that includes variants associated with many potential underlying disease states. These panels are used for diagnostic purposes.
Cancer Panels
Genetic panels for cancer can be of several types and may test for either germline or somatic variants. Their intended purpose can be for:
· Testing an asymptomatic patient to determine future risk of cancer
· Therapeutic testing of cancer cells from an affected individual to benefit the individual by directing targeted treatment based on specific somatic variants.
There are variations of panels for use in risk assessment or for directing targeted treatment. For our purposes, panels will be divided into the following types:
· Panels containing multiple variants indicating risk for a specific type of cancer or cancer syndrome (germline variants). These panels contain multiple related variants that indicate susceptibility to one or more cancers. They include germline variants and will generally be used for risk assessment in asymptomatic individuals who are at-risk for variants based on family history or other clinical data. An example of this type of panel would be one testing for multiple BRCA1 and BRCA2 variants associated with hereditary breast and ovarian cancer syndrome.
· Panels containing multiple variants associated with a wide variety of cancer types (somatic variants). These panels are generally used to direct treatment with drugs that target specific variants. They test for somatic variants from tissue samples of existing cancers. Many of these somatic variants are foundacross a wide variety of solid tumors. An example is the CancerNext Panel (Ambry Genetics), which tests for a broad number of somatic variants that can direct treatment.
Reproductive Panels
Reproductive panels test for variants associated with heritable conditions and are intended either for:
· Carrier testing of parent(s) preconception
· Carrier testing of parent(s) prenatal
· Prenatal (in utero) testing
Preconception testing usually tests for variants that are autosomal recessive or X-linked or, in some cases, for autosomal dominant variants with late clinical onset. Preconception tests can be performed on parents at-risk for a variant based on family history or can be done as screening tests in parents without a family history suggestive of a variant. Prenatal testing refers to tests performed during pregnancy. At present, prenatal testing for genetic variants is performed on the fetus, using amniocentesis or chorionic villous sampling. Testing of maternal blood for chromosomal aneuploidy is currently available, and in the future, it may be possible to test for fetal variants using maternal blood.
There are variations of panels for use in preconception or prenatal testing. For our purposes, panels will be divided into the following types:
· Panels containing variants associated with a single disorder. These panels are generally performed in at-risk individuals with a family history of a heritable disorder. An example of this type of panel would be a cystic fibrosis gene panel intended for use in individuals with a family history of cystic fibrosis.
· Panels containing variants associated with multiple disorders. These panels are generally performed as screening tests for parents without a family history of a heritable disorder. They can also be used to evaluate individuals with a family history of a heritable disorder. An example of this type of panel is the Signature Prenatal Microarray Panel.
Summary of Evidence
Genetic panels using next-generation technology or chromosomal microarray analysis are available for many clinical conditions. The major advantage of panels is the ability to analyze many genes simultaneously, potentially improving the breadth and efficiency of the genetic workup. A potential disadvantage of panels is that they provide a large of amount of ancillary information whose significance may be uncertain. Limited published evidence has reported that the analytic validity of panels approaches that of direct sequencing. The clinical validity and clinical utility of panels are condition-specific. The clinical validity of panels will reflect the clinical validity of the underlying individual variants. The clinical utility of panels will depend on the context in which they are used, ie, whether the advantages of panel testing outweigh the disadvantages for the specific condition under consideration.
SUPPLEMENTAL INFORMATION
Practice Guidelines and Position Statements
No guidelines or statements were identified.
U.S. Preventive Services Task Force Recommendations
Not applicable.
Ongoing and Unpublished Clinical Trials
A search of ClinicalTrials.gov in November 2017 did not identify any ongoing or unpublished trials that would likely influence this conceptual framework.
Appendix
Appendix Table 1. Categories of Genetic Testing
Category |
| 1. Testing of an affected individual’s germline to benefit the individual |
|
|
|
|
|
|
| 2. Testing cancer cells from an affected individual to benefit the individual |
|
|
|
|
|
|
| 3. Testing an asymptomatic individual to determine future risk of disease |
| 4. Testing of an affected individual’s germline to benefit family members |
| 5. Reproductive testing |
5a. Carrier testing: preconception
|
5b. Carrier testing: prenatal
|
5c. In utero testing: aneuploidy
|
5d. In utero testing: familial variants
|
5e. In utero testing: other
|
5f. Preimplantation testing with in vitro fertilization
|
Appendix Table 2. Criteria for Evaluating Panels by Type and Intent of Panel
Panel Category | Examples of Panels | Criteria for Evaluating Utility of Panel |
| 1. Diagnosis of hereditary, single-gene disorders |  | · All individual components of the panel have demonstrated clinical utility, OR test results that have not demonstrated clinical utility do not have a potential to cause harm
· Testing is performed in a CLIA-approved lab
· Analytic validity of panel approaches that of direct sequencing
· Panel testing offers substantial advantages in efficiency compared with sequential analysis of individual genes |
Category 1a – Diagnostic testing
Panels that include variants for a single condition | · Retinitis Pigmentosa Panel
· Leigh Disease Panel | · Includes all criteria for criterion 1 (Diagnosis of hereditary, single-gene disorders) |
Category 1b – Diagnostic testing
Panels that include variants for multiple conditions (indicated plus nonindicated conditions) | · Retinitis Pigmentosa/Leber Congenital Amaurosis Panel
· Noonan Syndrome and Related Disorders Panel | · Includes all criteria for criterion 1 (Diagnosis of hereditary, single-gene disorders) PLUS
· The impact of ancillary information is well-defined |
Category 1c – Diagnostic testing
Panels that include variants for multiple conditions (clinical syndrome for which clinical diagnosis not possible) | · X-linked Intellectual Disability Panel
· Marfan, Aneurysm and Related Disorders Panel
· Epilepsy Panel | · Includes all criteria for criterion 1 (Diagnosis of hereditary, single-gene disorders) PLUS
· The impact of ancillary information is well-defined
· Yield of testing is acceptable for the target population |
Category 1d – Risk Assessment
Risk assessment panels for at-risk individuals | · Most panels for hereditary conditions can be used for this purpose when there is not a known variant in the family | · Includes all criteria for criterion 1 (Diagnosis of hereditary, single-gene disorders) PLUS
· Yield of testing is acceptable for the target population |
| 2. Cancer panels |  | · All individual components of the panel have demonstrated clinical utility, OR test results that have not demonstrated clinical utility do not have a potential to cause harm
· Testing is performed in a CLIA-approved lab
· Analytic validity of panel approaches that of direct sequencing
· Panel testing offers substantial advantages in efficiency compared with sequential analysis of individual genes |
Category 2a – Risk assessment
Risk assessment panels for at-risk individuals | · Hereditary colon cancer syndromes panel
· BreastNext Panel | · Includes all criteria for criterion 2 (Cancer panels) PLUS
· Yield of testing is acceptable for the target population |
Category 2b – Targeted treatment based on variant analysis
· Panels with multiple variants intended to direct treatment – all indicated tests
· Effective targeted treatment based on variant analysis is available | · None identified | · Includes all criteria for criterion 2 (Cancer panels) PLUS
· Yield of testing is acceptable for the target population |
Category 2c – Targeted treatment based on variant analysis
· Panels with multiple variants intended to direct treatment (indicated plus nonindicated tests)
· Effective targeted treatment based on variant analysis has not been established | · CancerNext panels, when there is an effective targeted treatment for the specific type of cancer | · Includes all criteria for criterion 2 (Cancer panels) PLUS
· Impact of ancillary information is defined |
Category 2d
· Panels with multiple variants intended to direct treatment – no indicated tests for that particular cancer
· Effective targeted treatment based on variant analysis has not been established | · CancerNext panels, when there is no known effective treatment for the specific type of cancer | · Includes all criteria for criterion 2 (Cancer panels) PLUS
· Decision making based on potential results is defined
· Yield of testing is acceptable for the target population
· Impact of ancillary information is defined
· Probability that ancillary information leads to further testing or management changes |
| 3. Reproductive panels |  | · All individual components of the panel have demonstrated clinical utility, OR test results that have not demonstrated clinical utility do not have a potential to cause harm
· Testing is performed in a CLIA-approved lab
· Analytic validity of panel approaches that of direct sequencing
· Panel testing offers substantial advantages in efficiency compared with sequential analysis of individual genes |
Category 3a – Preconception testing of at-risk individuals
Panels that include only variants associated with increased risk | · Ashkenazi Jewish Carrier test Panel
· GoodStart Panel (customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Decision making based on genetic results is well-defined |
Category 3b - Preconception testing of at-risk individuals
Panels that include variants associated with increased risk plus other variants | · GoodStart Panel (full panel, not customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Decision making based on genetic results is well-defined
· Impact of ancillary information is defined |
Category 3c – Preconception screening
Panels intended for preconception testing – screening panels for different populations | · Counsyl Panel | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Yield of testing is acceptable for the target population
· Decision making based on genetic results is well-defined |
Category 3d – Prenatal screening
Panels that include only variants associated with increased risk | · Signature Prenatal Microarray Panel (customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Decision making based on genetic results is well-defined |
Category 3e - Prenatal screening
Panels that include variants associated with increased risk plus other variants | · Signature Prenatal Microarray Panel (full panel, not customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Yield of testing is acceptable for the target population
· Decision making based on genetic results is well-defined |
Category 3f – Preimplantation testing
Panels that include only variants associated with increased risk | · Signature Prenatal Microarray Panel (customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Decision making based on genetic results is well-defined |
Category 3g – Preimplantation testing
Panels that include variants associated with increased risk plus other variants | · Signature Prenatal Microarray Panel (full panel, not customized) | · Includes all criteria for criterion 3 (Reproductive panels) PLUS
· Yield of testing is acceptable for the target population
· Decision making based on genetic results is well-defined |
CLIA: Clinical Laboratory Improvement Amendments.
Appendix Figure 1. General Categories
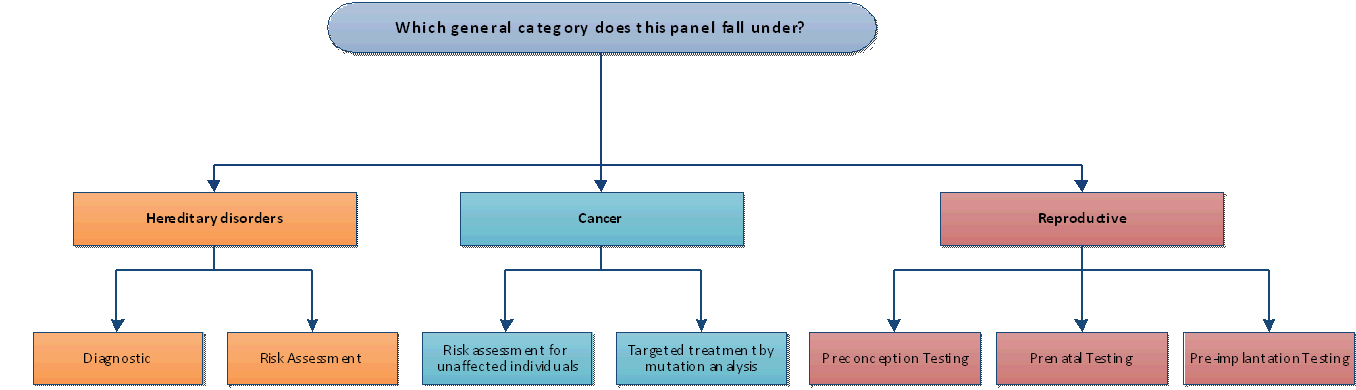
Appendix Figure 2. Algorithm for Evaluating the Utility for Hereditary Disease Panels

CLIA: Clinical Laboratory Improvement Amendments.
Appendix Figure 3. Algorithm for Evaluating the Utility of Cancer Panels
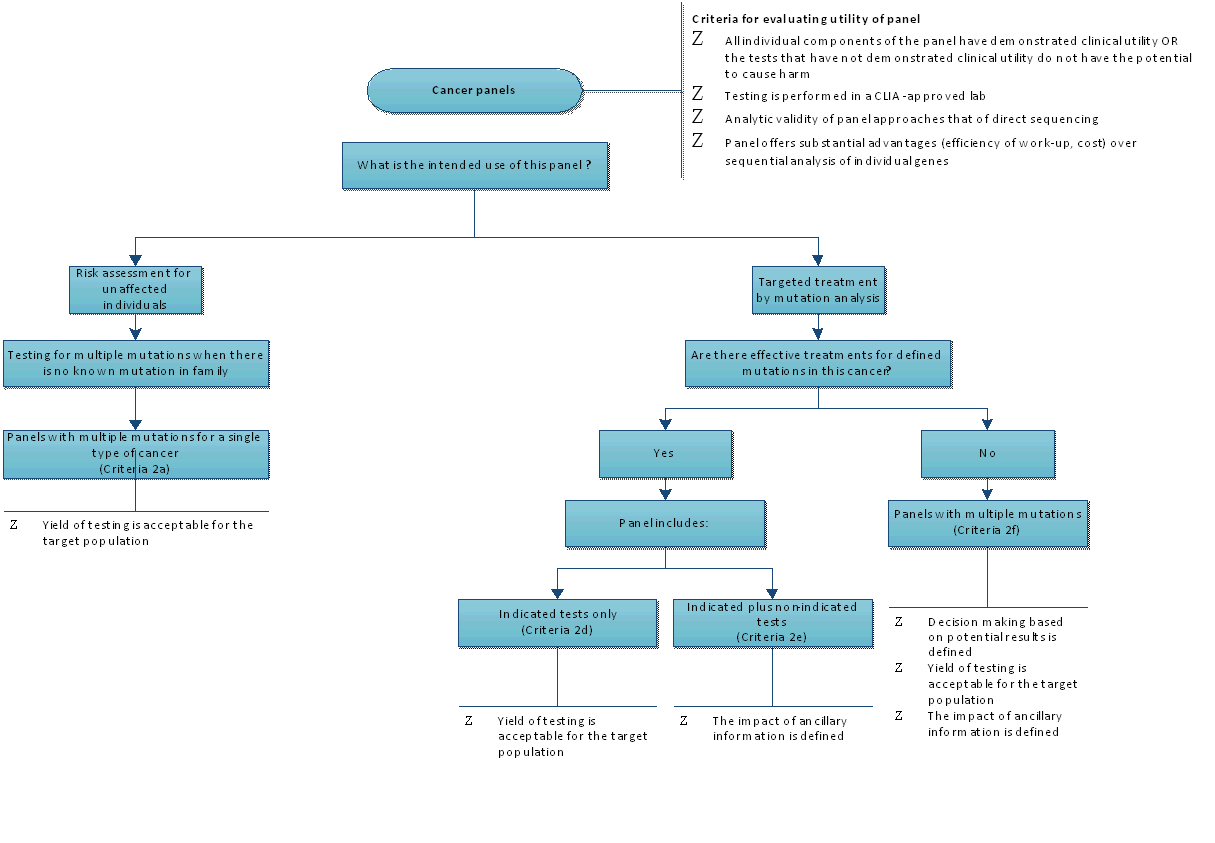
CLIA: Clinical Laboratory Improvement Amendments.
Appendix Figure 4. Algorithm for Evaluating Utility for Reproductive Panels
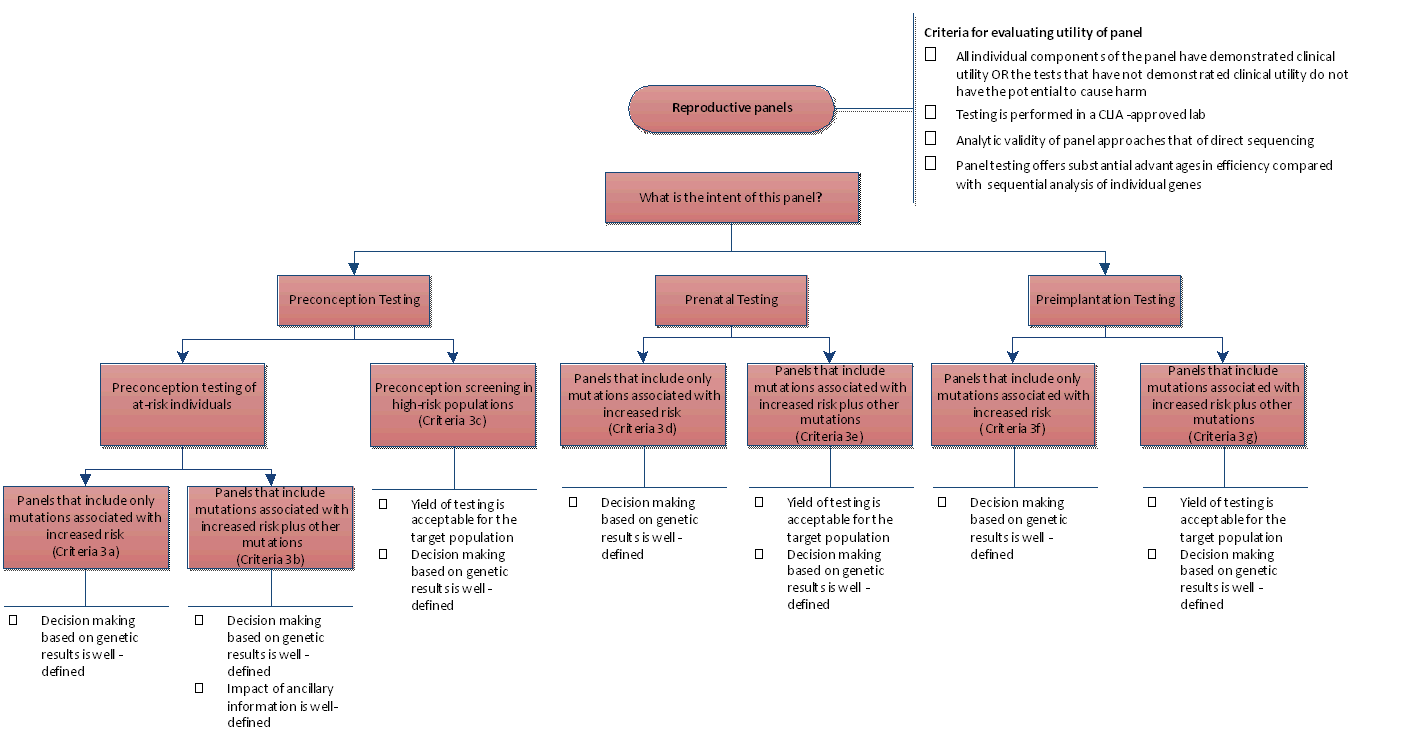
CLIA: Clinical Laboratory Improvement Amendments.]
________________________________________________________________________________________
Horizon BCBSNJ Medical Policy Development Process:
This Horizon BCBSNJ Medical Policy (the “Medical Policy”) has been developed by Horizon BCBSNJ’s Medical Policy Committee (the “Committee”) consistent with generally accepted standards of medical practice, and reflects Horizon BCBSNJ’s view of the subject health care services, supplies or procedures, and in what circumstances they are deemed to be medically necessary or experimental/ investigational in nature. This Medical Policy also considers whether and to what degree the subject health care services, supplies or procedures are clinically appropriate, in terms of type, frequency, extent, site and duration and if they are considered effective for the illnesses, injuries or diseases discussed. Where relevant, this Medical Policy considers whether the subject health care services, supplies or procedures are being requested primarily for the convenience of the covered person or the health care provider. It may also consider whether the services, supplies or procedures are more costly than an alternative service or sequence of services, supplies or procedures that are at least as likely to produce equivalent therapeutic or diagnostic results as to the diagnosis or treatment of the relevant illness, injury or disease. In reaching its conclusion regarding what it considers to be the generally accepted standards of medical practice, the Committee reviews and considers the following: all credible scientific evidence published in peer-reviewed medical literature generally recognized by the relevant medical community, physician and health care provider specialty society recommendations, the views of physicians and health care providers practicing in relevant clinical areas (including, but not limited to, the prevailing opinion within the appropriate specialty) and any other relevant factor as determined by applicable State and Federal laws and regulations.
___________________________________________________________________________________________________________________________
Index:
General Approach to Evaluating the Utility of Genetic Panels
Chromosomal Microarray Genetic Testing
Gene Test Panels
Next-Generation Sequencing
Genetic Panels
References:
1. Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. Nov 10 2009;106(45):19096-19101. PMID 19861545
2. Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. Jan 12 2011;3(65):65ra64. PMID 21228398
3. Foo JN, Liu J, Tan EK. Next-generation sequencing diagnostics for neurological diseases/disorders: from a clinical perspective. Hum Genet. Jul 2013;132(7):721-734. PMID 23525706
4. Lin X, Tang W, Ahmad S, et al. Applications of targeted gene capture and next-generation sequencing technologies in studies of human deafness and other genetic disabilities. Hear Res. Jun 2012;288(1-2):67-76. PMID 22269275
5. Raymond FL, Whittaker J, Jenkins L, et al. Molecular prenatal diagnosis: the impact of modern technologies. Prenat Diagn. Jul 2010;30(7):674-681. PMID 20572117
6. Simen BB, Yin L, Goswami CP, et al. Validation of a next-generation-sequencing cancer panel for use in the clinical laboratory. Arch Pathol Lab Med. Apr 2015;139(4):508-517. PMID 25356985
7. Yohe S, Hauge A, Bunjer K, et al. Clinical validation of targeted next-generation sequencing for inherited disorders. Arch Pathol Lab Med. Feb 2015;139(2):204-210. PMID 25611102
8. Sivakumaran TA, Husami A, Kissell D, et al. Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss. Otolaryngol Head Neck Surg. Jun 2013;148(6):1007-1016. PMID 23525850
9. Hiraki S, Rinella ES, Schnabel F, et al. Cancer risk assessment using genetic panel testing: considerations for clinical application. J Genet Couns. Aug 2014;23(4):604-617. PMID 24599651
10. Yorczyk A, Robinson LS, Ross TS. Use of panel tests in place of single gene tests in the cancer genetics clinic. Clin Genet. Sep 2015;88(3):278-282. PMID 25318351
11. EGL Genetics, Eurofins Clinical Diagnostics. Molecular Genetic Testing. 2017; http://www.egl-eurofins.com/tests/test-menu.php. Accessed November 30, 2017.
Codes:
(The list of codes is not intended to be all-inclusive and is included below for informational purposes only. Inclusion or exclusion of a procedure, diagnosis, drug or device code(s) does not constitute or imply authorization, certification, approval, offer of coverage or guarantee of payment.)
CPT*
HCPCS
* CPT copyright 2019 American Medical Association. All rights reserved. CPT is a registered trademark of the American Medical Association.
_________________________________________________________________________________________
Medical policies can be highly technical and are designed for use by the Horizon BCBSNJ professional staff in making coverage determinations. Members referring to this policy should discuss it with their treating physician, and should refer to their specific benefit plan for the terms, conditions, limitations and exclusions of their coverage.
The Horizon BCBSNJ Medical Policy Manual is proprietary. It is to be used only as authorized by Horizon BCBSNJ and its affiliates. The contents of this Medical Policy are not to be copied, reproduced or circulated to other parties without the express written consent of Horizon BCBSNJ. The contents of this Medical Policy may be updated or changed without notice, unless otherwise required by law and/or regulation. However, benefit determinations are made in the context of medical policies existing at the time of the decision and are not subject to later revision as the result of a change in medical policy
____________________________________________________________________________________________________________________________ |